Biomolecular Science
Maho Yagi(Assis. Prof.) Saeko Yanaka(Assis. Prof.)
RESEARCH THEMES
Elucidation of the Mechanisms Underlying Dynamical Ordering of Biomolecular Systems for Creation of Integrated Functions
Living systems are characterized as dynamic processes of assembly and disassembly of various biomolecules that are self-organized, interacting with the external environment. The omics-based approaches developed in recent decades have provided comprehensive information regarding biomolecules as parts of living organisms. However, fundamental questions still remain unsolved as to how these biomolecules are ordered autonomously to form flexible and robust systems. We develop multidisciplinary research area, including fields of biology, chemistry, physics, and aims to elucidate the mechanisms underlying the dynamic ordering of biomolecules for the creation of integrated functions.
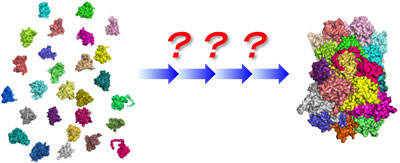 |
| Formation of supramolecular machinery through dynamic assembly and disassembly of biomolecules |
SELECTED PUBLICATIONS
- T. SUZUKI, M. KAJINO, S. YANAKA,T. ZHU,H. YAGI,T. SATOH,T. YAMAGUCHI and K. KATO, "Conformational analysis of a high-mannose-type oligosaccharide displaying glucosyl determinant recognised by molecular chaperones using NMR-validated molecular dynamics simulation,"ChemBioChem 18, 396-410 (2017).
- G. YAN, T. YAMAGUCHI, T. SUZUKI, S. YANAKA, S. SATO, M. FUJITA and K. KATO, "Hyper-assembly of self-assembled glycoclusters mediated by specific carbohydrate?carbohydrate interactions," Chem. Asian J. 12, 968-972 (2017).
- R. YOGO, S. YANAKA, H. YAGI, A. MARTEL, L. PORCAR, Y. UEKI, R. INOUE, N. SATO, M. SUGIYAMA and K. KATO, "Characterization of conformational deformation-coupled interaction between immunoglobulin G1 Fc glycoprotein and a low-affinity Fcγ receptor by deuteration-assisted small-angle neutron scattering," Biochem. Biophys. Rep. 12, 1-4 (2017).
- T. SATOH, C. SONG, T. ZHU, T. TOSHIMORI, K. MURATA, Y. HAYASHI, H. KAMIKUBO, T. UCHIHASHI and K. KATO, "Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT," Sci. Rep. 7, Article number: 12142 (2017).
- S. YANAKA, T. YAMAZAKI, R. YOGO, M. NODA, S. UCHIYAMA, H. YAGI and K.KATO, "NMR detection of semi-specific antibody interactions in serum environments," Molecules 22, Article number: 1619 (2017).
- Y. SAKAE, T. SATOH, H. YAGI, S. YANAKA, T. YAMAGUCHI, Y. ISODA, S. IIDA, Y. OKAMOTO and K. KATO, "Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fcγ receptor IIIa," Sci. Rep. 7, Article number: 13780 (2017).


